Protein Structures - Cn3D
The application Cn3D will open enabling you to see the structure of your protein. You can rotate the 3-D structure by dragging it with your mouse. The catalytic active region is shown in red.
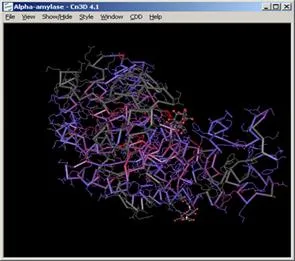
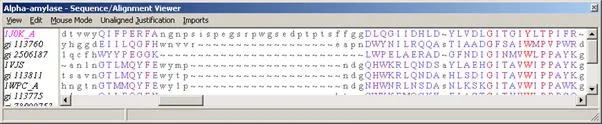
The color key matches the amino acid sequence information in the window that appears below the 3-D representation of your protein:
Change the display format of Cn3D by selecting Style –> Rendering Shortcuts –> Worms:
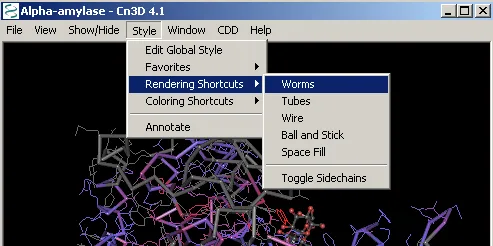
Now you should be able to rotate the structure to clearly see the α/β barrel site in the center.
Protein Structures: Comparisons
Now that you know what the catalytic site looks like, you can search for the 3-D structure of the enzymes used in this lab and see how they compare.
- Close the CDD windows and return to the main NCBI website by clicking the NCBI logo in the upper left corner.
- Click on STRUCTURE at the top of the page.
- At the Search Entrez Structure, enter “human alpha amylase” and click GO.
- On the results list, find and select Human Salivary Amylase (1SMD). Click on VIEW 3D STRUCTURE.
- Rotate the model of the enzyme – can you see the characteristic catalytic site? This site does not show the catalytic site in red, but you can highlight a section of the sequence in the lower window, and it will also be highlighted on the model.
- Minimize the 3D model, and go back one page. Unfortunately, there are no structure models for either corn or clams in the database, but there is one for barley. (Before viewing the structure of the barley enzyme, return to the ClustalW page and compare the barley and corn sequences to determine if this subsitute is valid). Enter “barley alpha amylase” and click GO. On the results list, choose the crystal structure for barley alpha amylase (1RPK). Click on VIEW 3D STRUCTURE
- Rotate the model of the enzyme – can you see the characteristic catalytic site? Maximize the window with the human enzyme model and compare the two side by side.