DNA and RNA structure: nucleic acids as genetic material
Last month, we covered the definition of molecular diagnostics (MDx) as the detection of nucleic acids to provide clinical information. The methods used to do so, as well as the intrinsic biological roles of DNA and RNA in the storage and transmission of an organism’s genetic information, turn out to be intimately related to their physical structures. As a precursor to understanding MDx methods we must therefore turn our attention this month to DNA and RNA structure.

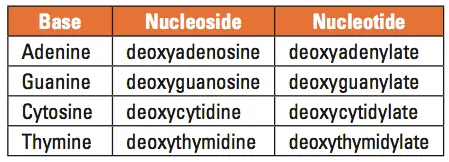
Table 1. Names of the DNA bases, nucleosides, and nucleotides
DNA is best considered in terms of its constituent subparts. The reader is probably aware that there are four such subparts, and that each of the four consists of a heterocyclic nitrogen-containing molecule (the nitrogenous base, or just “base,” in context); the base covalently linked to a 5 carbon sugar (2′ – deoxyribose), with the base plus sugar being referred to as a “nucleoside”; and the nucleoside with the covalent addition of one, two, or three phosphate groups to the 5′ carbon, which respectively make up a nucleoside mono-, di-, or triphosphate (collectively referred to as a “nucleotide”). Table 1 gives the names for each of these 12 molecules, and Figure 1 shows the structures for all four bases, plus an example of deoxyadenylate with the base, nucleoside, and nucleotide portions labelled. For the other nucleosides and nucleotides, the reader can just envision exchanging the base part of the structure.
We’ve been proper here and included the “deoxy” part of each name, which signifies that it’s a DNA constituent with a 2′-deoxyribose sugar that we’re discussing. In common usage, sometimes this “deoxy” is just abbreviated as “d-,”or often, if the context is clearly discussing DNA, left out altogether.
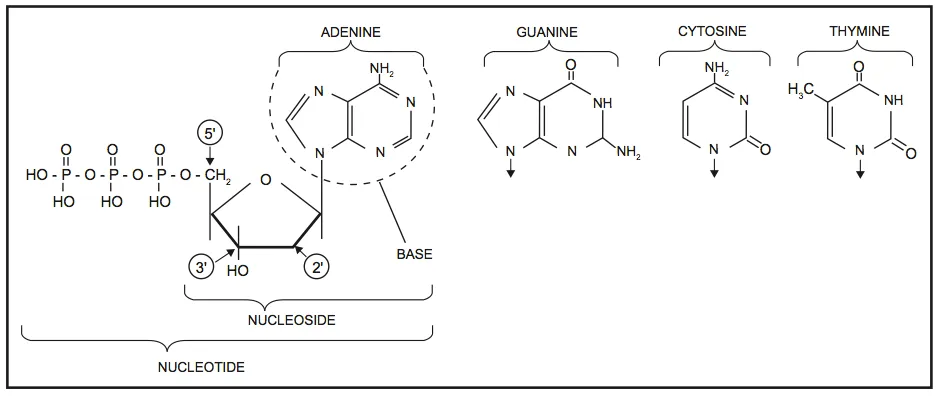
Figure 1. Structures of the bases, with adenine/adenosine/adenylate shown as an example of nucleoside and nucleotide forms
Consider Figure 1. First, on the deoxyribose portion, carbons labelled 2′ (pronounced “2 prime”), 3′, and 5′ have been labelled. The 2′ position here doesn’t have a sugar (thus it’s 2′-deoxyribose), the reactive parts of the molecule are the 3′ –OH, and the phosphate groups are attached to the 5′ position. We’ll come back to these later. Second, look at the bases, and notice the –NH, N, and =O groups either in the ring or exocyclic (sticking out). These form a set of H-bond donors and acceptors, as discussed last month. It’s important to note that each base has its own unique pattern of these. Note also that two of the bases (adenine and guanine) are large, two-ring structures (purines), while cytosine and thymine are smaller, single-ring entities (pyrimidines).
A DNA molecule is initially composed of a string of nucleoside monophosphates (NMPs) held together by covalent bonds between the 3′ –OH of one subunit, and the innermost (alpha) phosphate group on the 5′ end of another NMP (the other two phosphate groups are released in the joining reaction). The 3′ end of this NMP is then attached to the 5′ alpha phosphate of another NMP, and so on, to create a long, single stranded chain of NMPs. Together the sugar and phosphate parts form a repeating backbone of this strand, with the bases projecting off to the side. The strand still has exactly one free and reactive 5′ end and at the opposite end, a free, reactive 3′ –OH group. This is known as the polarity of the strand, and by convention we consider single DNA strands from their 5′ towards their 3′ ends. As the backbone is exactly the same repeating unit, the NMPs can be strung together in any order, and we generally refer to this order by just naming the 5′ to 3′ order of the base components such as 5′-ACTGAATGCA-3′ or 5′-TGGATC-3′. Chains of roughly 6 to 50 or so joined nucleotides are generically called oligonucleotides, while longer chains are polynucleotides.
It is in such polynucleotides that DNA gains its biological utility. If there are two strands of DNA and they orient their polarities in opposite directions (antiparallel) to each other, their bases can interact and H-bond to each other—but only under one strict rule. An adenine on one strand can line up its H-bond donors and acceptors with a thymine on the opposite strand, forming a total of 2 H bonds; and a guanine can line up similarly with a cytosine, forming 3 H-bonds. This specific pairing is referred to as the sequences or strands being complementary. You’ll note that each base pair consists of a purine and a pyrimidine, such that the spacing between backbones is the same on an A:T base pair and a G:C base pair. This regularity along two long antiparallel strands leads to the famous, repeating “double helix” structure of DNA familiar to everyone. Because of complementarity, knowledge of the sequence of bases on one strand automatically defines for us the sequence of bases on the antiparallel second strand. For instance, from our examples in the paragraph above, the antiparallel strands would have to be 5′-TGCATTCAGT-3′ and 5′-GATCCA-3′. (If the reader is unsure of this, try writing these out on a piece of paper, with one sequence above the other and the 5′ ends of each strand at opposite ends; then notice how the A:T and G:C pairing rules are always observed).
What holds the two strands of the double helix together? Recall last month’s “not-really-accurate-but-mentally-helpful” analogy of H-bonds as being like tiny magnets. When two DNA single strands can align with each other, each of the H-bonds that can form between the correct A:T and G:C base pairs has a small attractive energy. Individually, each H-bond formed is quite weak, but when a number of them occur in series such as between two complementary strands, the effect is additive and the result is a non-covalent but strong net force holding the strands together. If the strands are not complementary, then the H-bond donors and acceptors don’t line up, and this collective attractive force isn’t present to hold them together. Additionally, if two non-complementary strands try to pair up any two purines, they’re physically too large to both fit between the backbones; and if two pyrimidines are placed across from each other, they’re too small and their respective H-bond donors and acceptors not only don’t match up, but are also too far apart to effectively interact. Only a purine—pyrimidine pair has the right size, and only A:T and G:C sets match each other’s H-bonding capacities.
Asking what holds two strands of DNA together presupposes there’s some other force trying to pull the strands apart. There is, and that force is none other than heat. Heat, after all, is just random motion of molecules, and the force of this vibrational motion depends on temperature. At physiological temperature ranges, the thermal energy in any one single nucleotide is more than the energy it can find in its complementary H-bonds. Thus, a free adenylate in a cell doesn’t automatically pair up with the first thymidylate it passes; their vibrational energies carry them apart as they surpass their net attractive forces. As two complementary strands get longer, the additive nature of the H-bond energies comes into play, and at some total number of potential H-bonds between two single DNA strands, their energy surpasses the temperature dependent vibrational energies, and the two strands stick together. What this exact temperature is depends on the length of the DNA molecules (longer chains of nucleotides have more bases and thus more potential H-bonds) and also the sequence; as mentioned above an A:T base pair has two H-bonds while a G:C base pair has three, so sequences with more “G:C content” stick together more strongly than sequences with more A:T base pairs.
The process whereby complementary DNA strands find each other and pair up their H-bonds to effectively overcome thermal energies trying to shake them apart is referred to as annealing, and two DNA strands H-bonded to each other are referred to as annealed. The attractive force between two annealed strands is a fixed value based on total number of H-bonds; the thermal energy acting to try to vibrate the strands apart of course is not fixed, and can be manipulated by heating or cooling of the environment. If the environment is heated to the point where thermal energy overcomes the attractive force between two DNA strands, vibration wins out and the strands separate or denature. The temperature at which thermal energy exactly matches H-bond energy for a given DNA paired sequence, and the strands hover between annealing and denaturation, is known as the melting temperature or Tm of the sequence. To get a sense of the energy and temperature ranges here, a DNA sequence of about equal numbers of A:T and G:C base pairs, about 13 base pairs in total length, has a Tm of around 37°C, while a similar random sequence of about 20 bases has a Tm of about 51°C. Making sequences longer or more G:C rich raises the Tm, but there is a limit; any DNA sequence will effectively denature as its environment approaches 100°C and the aqueous environment starts a liquid-to-gas phase change. In fact, since the solvent isn’t pure water, the limit temperature at which essentially all DNA sequences denature is slightly lower, in the range of 95°C. We’ll see later how MDx techniques utilize the processes of annealing and denaturation by temperature manipulation as the core of multiple methods.
We’ll close off this month’s section with a brief consideration of ribonucleic acid (RNA). While chemically similar to DNA, it has some critical differences which can be briefly summarized:
The majority of MDx focuses on DNA as a target, and so will we. However, RNA can also be a useful MDx target and where this is so, we’ll generally approach the subject as we have here, with a detailed discussion of the DNA case, and then a brief summary of how this differs for RNA.