Inverse PCR:
Here, the unknown DNA sequence adjacent to the known template DNA is amplified using the primers which amplify the flanking (unknown) region near the know DNA template.
Quite difficult to understand?
No problem, the present article is only about the inverse PCR. I know all other resources available online about the inverse PCR are confusing.
In this present article, we will understand the mechanism of inverse PCR and its application in molecular genetics.
I bet you read the article till the end, you will understand the fundamental basics behind the inverse PCR.
PCR is a technique in which the DNA is amplified using a set of the sequence-specific complementary primers in the enzymatic cyclic temperature dependent reaction.
Inverse PCR is just a variant of the conventional PCR. The inverse PCR method is originally developed by Howard Ochman and coworker in the year 1988.
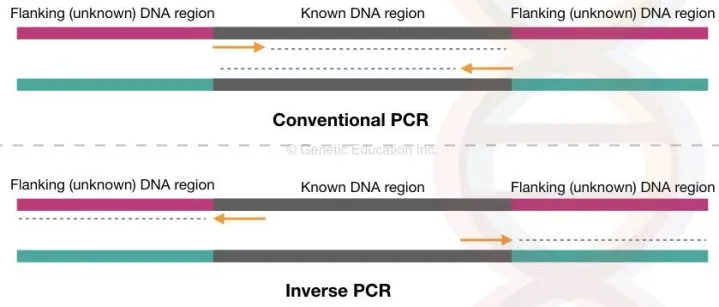
Firstly we have to understand the basic difference between conventional PCR and the inverse PCR.
Now see the figure, in the conventional PCR, our target DNA is known to us, we have the sequence information of that.

depending upon that the primers are designed to amplify that know complementary DNA sequence. On two different single-stranded DNA, primers are bind and synthesised towards each other.
While in the inverse PCR, the region of the DNA is unknown. We do not have any information about what types of DNA is inserted into the genome.
However, we design primers complementary to the known DNA region but instead of extending towards each other, the primers extend DNA away from each other. See the above figure.
Although the Taq DNA polymerase is the same in both types of PCR. No special DNA polymerase is required in the inverse PCR.
With the help of the sequence information of known DNA region, the unknown flanking region of the DNA or the inserted DNA is amplified into the cyclic enzymatic reaction using the known DNA sequence-specific primers. DNA synthesis occurs outside the known DNA region.
Restriction digestion is performed on the genomic DNA which digests only the unknown flanking regions but not the known DNA region.
ligating the flanking regions of unknown DNA generates circular DNA which is amplified using the set of primers.
The entire process of inverse PCR is divided into 5 steps:
1. Identification of known DNA region having flanking unknown DNA sequence.
2. Restriction digestion of gDNA
3. ligation of digested unknow DNA fragments
4. Amplification of ligated circular DNA molecule
5. Sequencing of the unknown DNA region

1. Identification of known DNA region having flanking unknown DNA sequence:
In the very first step, we have to identify the target. Inverse PCR is used for identification gene rearrangements, transposons and jumping gene studies. By identifying the known location near the unknown DNA region, design the primers complementary to the known DNA region helps to amplify the unknown region.
In the next step, extract DNA using any of the DNA extraction methods listed below,
1. Different types of DNA extraction methods
2. Phenol-chloroform DNA extraction method
3. CTAB DNA extraction method
4. Proteinase K DNA extraction method
After DNA extraction, quantify the DNA and check the purity of the DNA because the purity matters a lot in the amplification.
Store the sample in TE buffer at 4°C.
2. Restriction digestion of gDNA:
Restriction digestion is a process in which the DNA having the restriction site is digested using the known restriction endonuclease.
For more detail on the restriction digestion read our article: What is Restriction Digestion and how to do it?
Now, digest the gDNA using a desired restriction digestion endonuclease.
Choose the endonuclease which can only cut the unknown flanking regions but not the known DNA regions.
If you doing it for higher eukaryotes, thousands of digested DNA fragments may be generated.

3. Ligation of digested unknow DNA fragments:
The sticky DNA ends generated during restriction digestion are now ligated, using the DNA ligation assay.
Ligation is a process in which the ends of the DNA are joined using physical means or by an enzyme. Ligase is the enzyme used to ligate two DNA molecules.
Here instead of enzyme-mediated ligation, The DNA is self-ligated using the intermolecular ligation. For that low concentration of digested DNA is used.
If the concentration of digested DNA is too high, it might hinder in the ligation. Hence use low concentration digested DNA.
We can also use ligation assay using the ligase which is more preferable.
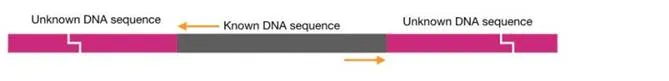
Now the ligated ends of DNA generates a circular DNA molecule shown into the figure below,

4. Amplification of ligated circular DNA molecule:
The template is ready for the amplification. Here we can amplify the single template or both, it depends on us.
The unknown DNA fragment is now present between the two known fragments.
On the one side of the circular DNA is our known DNA sequence for what we had design primers. See the figure, the primers bind to the known DNA sequence but amplifies outward to the known DNA.
Interestingly, here the 3′ OH end of the primers are outward to the known DNA sequence.
With the help of the Taq DNA polymerase, the unknown region of the DNA is amplified.

5. Sequencing of the unknown DNA region:
Remember here we are doing amplification for obtaining a large amount of the unknown DNA amplicon, which type of sequence is inserted to the DNA is still unknown to us. For that, we have to do DNA sequencing.
After the inverse PCR, the amplicons are sent for DNA sequencing at where the nucleotide sequence of unknown DNA between the two know DNA region is determined.

Once it is sequenced, the sequence is cross-checked with other DNA sequences or with other genomes for checking the duplication, translocation or insertion.
The detail application of this technique will be discussed in the application section.