Dynamic programming and sequence alignment
Genetics and string algorithms
Strands of genetic material — DNA and RNA — are sequences of small units called nucleotides. For purposes of answering some important research questions, genetic strings are equivalent to computer science strings — that is, they can be thought of as simply sequences of characters, ignoring their physical and chemical properties. (Although, strictly speaking, their chemical properties are usually coded as parameters to the string algorithms you’ll be looking at in this article.)
This article’s examples use DNA, which consists of two strands of adenine (A), cytosine (C), thymine (T), and guanine (G) nucleotides. DNA’s two strands are reverse complements of each other. A and T are complementary bases, and C and G are complementary bases. This means that A s in one strand are paired with T s in the other strand (and vice versa), and C s in one strand are paired with G s in the other strand (and vice versa). So, if you know the sequence of one strand’s A s, C s, T s, and G s, you can derive the other strand’s sequence. Hence, you can think of a DNA strand simply as a string of the letters A, C, G, and T.
Dynamic programming
Dynamic programming is an algorithmic technique used commonly in sequence analysis. Dynamic programming is used when recursion could be used but would be inefficient because it would repeatedly solve the same subproblems. For example, consider the Fibonacci sequence: 0, 1, 1, 2, 3, 5, 8, 13, … The first and second Fibonacci numbers are defined to be 0 and 1, respectively. The _n_th Fibonacci number is defined to be the sum of the two preceding Fibonacci numbers. So, you can calculate the _n_th Fibonacci number with the recursive function in Listing 1:
Listing 1. Recursive function for calculating nth Fibonacci number
public int fibonacci1(int n) {
if (n == 0) {
return 0;
} else if (n == 1) {
return 1;
} else {
return fibonacci1(n ‑ 1) + fibonacci1(n ‑ 2);
}
}
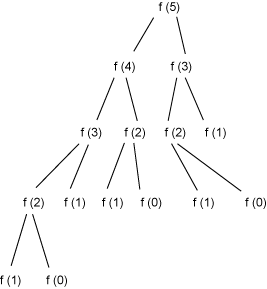
But Listing 1’s code is inefficient because it solves some of the same recursive subproblems repeatedly. For example, consider the computation of fibonacci1(5), represented in Figure 1:
Figure 1. Recursive computation of Fibonacci numbers

In Figure 1 you can see, for example, that fibonacci1(2) is computed three times. It would be much more efficient to build the Fibonacci numbers from the bottom up, as shown in Listing 2, rather than from the top down:
Listing 2. Building Fibonacci numbers from the bottom up
public int fibonacci2(int n) {
int[] table = new int[n + 1];
for (int i = 0; i < table.length; i++) {
if (i == 0) {
table[i] = 0;
} else if (i == 1) {
table[i] = 1;
} else {
table[i] = table[i ‑ 2] + table[i ‑ 1];
}
}
return table[n];
}
Listing 2 stores the intermediate results in a table so that you can reuse them, rather than throwing them away and computing them multiple times. It’s true that storing the table is memory-inefficient because you use only two entries of the table at a time, but ignore that fact for now. I’m doing it this way to motivate your use of similar tables (although they will be two-dimensional) in this article’s more complicated later examples. The point is that Listing 2’s implementation is much more time-efficient than Listing 1’s. Listing 2’s implementation runs in O(n) time. I won’t prove this, but the running time of Listing 1’s naive, recursive implementation is exponential in n.
This is exactly how dynamic programming works. You take a problem that could be solved recursively from the top down and solve it iteratively from the bottom up instead. You store your intermediate results in a table for later use; otherwise, you would end up computing them repeatedly — an inefficient algorithm. But dynamic programming is usually applied to optimization problems like the rest of this article’s examples, rather than to problems like the Fibonacci problem. The next example is a string algorithm, like those commonly used in computational biology.